

Next: Conclusion
Up: Segmentation of Brain MR
Previous: Experiments
We note from the above experiments that using the HMRF-EM
algorithm, not only does the segmentations improve significantly
but the bias field estimates do too, although the HMRF-EM
algorithm itself is not directly related to bias field estimation.
This is due to the fact the bias field estimation relies strongly
on the classification.
A practical issue has to be addressed in the 3D implementation of
the HMRF-EM algorithm. Theoretically the MRF neighbourhood system
should be 3-dimensionally isotropic. However, the slice thickness
of a 3D volume is often larger than the intra-slice voxel
dimensions. In such a situation, an isotropic neighbourhood system
may cause problems. Therefore an anisotropic 3D neighbourhood
system is used with a smaller weight across slices.
Although the HMRF-EM framework itself is theoretically sound, the
initial estimation based on thresholding is rather heuristic. Due
to the high variability of brain MR images in terms of their
intensity ranges and contrasts between brain tissues, it is not
guaranteed that the thresholding procedure will produce perfect
results. In most cases, however, the final segmentation results
are stable even with slightly different initial estimates. This is
largely attributable to the robust HMRF-EM algorithm. However, as
a local minimization method, the EM algorithm can be trapped in a
local minimum. In some cases, where the image is poorly-defined,
the thresholding procedure may fail to find the right thresholds
for brain tissues, especially the threshold for GM and WM. With an
initial condition far from normal, the EM procedure is likely to
give a wrong final segmentation. In general, the initial
estimation is a difficult problem and it will certainly be an
important issue to be studied in future work.
With respect to the computational load, the whole algorithm is
slightly slower than the original FM model-based MEM algorithm due
to the additional MRF-MAP classification and the EM fitting
procedure. However, by employing the fast deterministic ICM method
and certain optimizations to the program, it runs reasonably
quickly. Currently, it takes 16 seconds for a
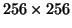 2D
image and about 10 minutes for a 3D volume data with 40
2D
image and about 10 minutes for a 3D volume data with 40
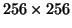 slices in an Intel PII 400MHZ-based system.
slices in an Intel PII 400MHZ-based system.


Next: Conclusion
Up: Segmentation of Brain MR
Previous: Experiments
Yongyue Zhang
2000-05-11