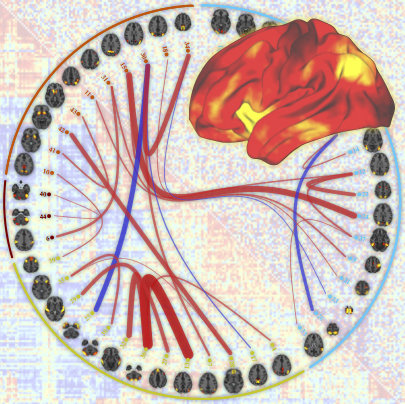
A positive-negative mode of population covariation links
brain connectivity, demographics and behavior
Stephen M Smith,
Thomas E Nichols, Diego Vidaurre, Anderson M Winkler, Timothy E J
Behrens, Matthew F Glasser, Kamil Ugurbil, Deanna M Barch, David C
Van Essen & Karla L Miller
Nature
Neuroscience, early
online, 2015
Please send feedback to steve@fmrib.ox.ac.uk
The netmats fed into the CCA were from the HCP500 "PTN" (Parcels, node Timeseries and Netmats) data release available here. We primarily used the 200-dimensional group-ICA parcellation, with node-timeseries estimation method "ts2" (multiple regression of voxelwise-timeseries data against ICA spatial maps) and the partial correlation netmats.
The subject measures (SMs) also fed into the CCA combine data from the two HCP files referred to as "Behavioral data" and "Restricted data". We cannot reproduce those here, because it is necessary to sign the HCP access forms to get this subject-sensitive data, and access the data files via the HCP website (you may need to change the near-the-top drop-down menu to "sensitive", and then can download the "Behavioral" and "Restricted" data files).
To try to make it as easy as possible to generate the appropriate data matrix from these two HCP data files (to match what was fed into our CCA), we include here the file column_headers.txt, which describes how to organise the columns when generating the "vars" subjectsXsubject-measures matrix used in our CCA (see code below). This column_headers file has one text string per row; each entry corresponds to one column in "vars", and matches the appropriate column heading in the two original HCP data files described above.
Other information needed is:
A) The "rfMRI_motion" summary head
motion file, including subject IDs,
is here (column 2 of this needs to
become column 7 in "vars.txt").
B) The "quarter/release"
information about recon-code-version
is here, with the rows ordered according
to the same set of subject IDs as in the head motion file (this
needs to become column 2 in "vars.txt").
You can download the basic CCA code used in our paper here.
We have rerun the same analysis using the HCP900 (820 complete
rfMRI subjects) PTN release and obtained a very similar result
(subject-measure weights correlate with r=0.94). You can download a
matlab workspace file with the (HCP900 ICA200) netmat CCA weights and
subject-measure weights (matching the list of variables linked to
above) here.