

Next: Determining Basis Set Constraints
Up: Model
Previous: Constraining Basis Function Linear
Choosing a Basis Set
Basis sets used previously range from a single canonical HRF plus
its temporal derivative to a set of Gamma
functions Friston et al. (1998a)). These basis functions are then
separately convolved with the known stimulus to give the same
number of regressors as there are basis functions for use in the
linear model.
Hossein-Zadeh and Ardekani (2002) and Friman et al. (2003) have previously shown how we
can generate a basis set using singular value decomposition (SVD).
This produces a basis set from samples of the HRF or regressors
resulting from a parametric forward model of the haemodynamics.
This is the approach we take in this paper.
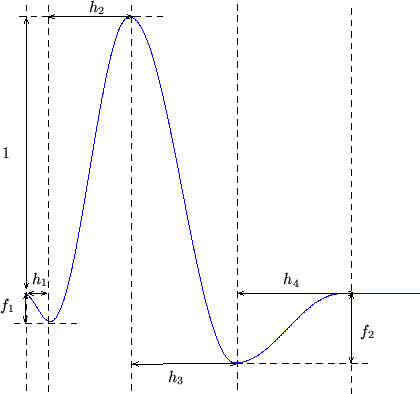
In this paper we base the basis set on a parameterised model of
the HRF. The HRF is parameterised by four half-cosines, requiring
six parameters, as illustrated in
figure 1.
Figure 1:
Parameterisation of the HRF into four half-period
cosines. There are six parameters.
 |
To complete the HRF parameterised model we need to specify
probabilities for the parameter values in the HRF model from which
we generate physiologically plausible HRF shapes. With this
information we can then draw HRF samples.
Figure 2 shows 20 HRF samples drawn from
the half-cosine parameterisation using the HRF parameter value
probabilities:
 |
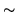 |
Uniform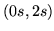 |
|
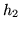 |
 |
Uniform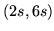 |
|
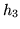 |
 |
Uniform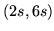 |
|
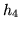 |
 |
Uniform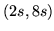 |
|
 |
 |
0 |
|
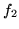 |
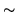 |
Uniform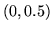 |
(14) |
We strongly emphasise that this is one of many possible choices of
obtaining HRF samples. An alternative, attractive approach would
be to use a physiologically based model, such as the Balloon
model (Friston et al., 2000), with physiologically meaningful
parameters which can be given sensible ranges.
Using a probabilistic model of choice, one can obtain a set of
samples of the HRF, which represents the space of HRFs expected.
The 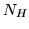 HRF samples, each of length
HRF samples, each of length
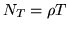 , can be placed
into a
, can be placed
into a
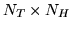 matrix,
matrix, 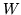 . To obtain a basis set that
spans this space of HRFs we perform a Singular Value Decomposition
(SVD) on this matrix. This gives us
. To obtain a basis set that
spans this space of HRFs we perform a Singular Value Decomposition
(SVD) on this matrix. This gives us 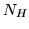 eigenHRFs (of length
eigenHRFs (of length
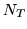 ) and
) and 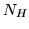 corresponding eigenvalues (describing the power
each corresponding eigenHRF explains).
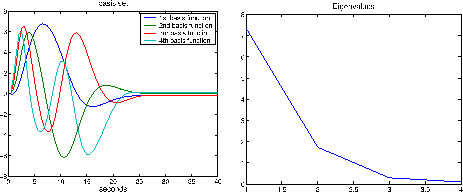
Figure 3 shows the four eigenHRFs (and their
corresponding eigenvalues) with the largest eigenvalues obtained
from an SVD on
corresponding eigenvalues (describing the power
each corresponding eigenHRF explains).
Figure 3 shows the four eigenHRFs (and their
corresponding eigenvalues) with the largest eigenvalues obtained
from an SVD on 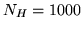 HRF samples of length
HRF samples of length 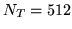 (resolution of
(resolution of 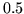 seconds) from the half-cosine
parameterisation. It is worth noting that the first three
eigenHRFs/basis functions look remarkably like the commonly used
canonical, delay (temporal) derivative and width (dispersion)
derivative of a Gamma/Gaussian parameterised HRF (in that order).
To form our basis set we need to decide how many of the largest
eigenHRFs we want to include. For the rest of this paper we use
the
seconds) from the half-cosine
parameterisation. It is worth noting that the first three
eigenHRFs/basis functions look remarkably like the commonly used
canonical, delay (temporal) derivative and width (dispersion)
derivative of a Gamma/Gaussian parameterised HRF (in that order).
To form our basis set we need to decide how many of the largest
eigenHRFs we want to include. For the rest of this paper we use
the 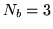 largest eigenHRFs as our basis set.
largest eigenHRFs as our basis set.
Figure 2:
20
HRF samples drawn from the half-cosine parameterisation
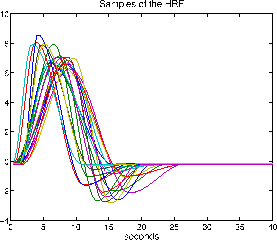 |
Figure 3:
Four eigenHRFs (and their corresponding eigenvalues) with
the largest eigenvalues from 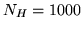 HRF samples of length
HRF samples of length
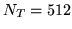 (resolution of
(resolution of 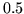 seconds) from the half-cosine
parameterisation.
seconds) from the half-cosine
parameterisation.
 |


Next: Determining Basis Set Constraints
Up: Model
Previous: Constraining Basis Function Linear

