We have a number of choices for how we use the posterior
distribution
![]() . We could simply use the posterior,
. We could simply use the posterior,
![]() , to build up posterior probability maps
representing the probability of activation at each
voxel (10). Another possibility is the use of
(spatial) mixture
modelling (14,23,6) to classify
voxels as activating and non-activating. We do not attempt to
explore or discuss the relative merits of these approaches in this
paper. Here, we consider another possibility of the inference
produced if we mimic null-hypothesis frequentist inference (i.e.
controlling a False Positive Rate (FPR)) by assuming that under
the null hypothesis the z-statistics, that the fully Bayesian
[BIDET] approach produces, are standardised (zero mean and
standard deviation of one) Normally distributed.
, to build up posterior probability maps
representing the probability of activation at each
voxel (10). Another possibility is the use of
(spatial) mixture
modelling (14,23,6) to classify
voxels as activating and non-activating. We do not attempt to
explore or discuss the relative merits of these approaches in this
paper. Here, we consider another possibility of the inference
produced if we mimic null-hypothesis frequentist inference (i.e.
controlling a False Positive Rate (FPR)) by assuming that under
the null hypothesis the z-statistics, that the fully Bayesian
[BIDET] approach produces, are standardised (zero mean and
standard deviation of one) Normally distributed.
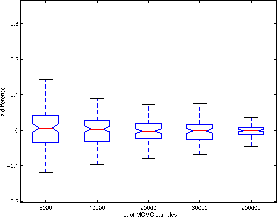
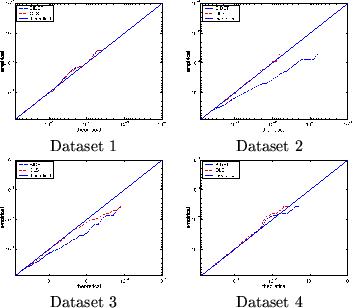
To examine this possibility, figure 6 shows the log probability-log probability plots for the four different datasets for [BIDET] and [OLS]. These are plots of the nominal/theoretical frequentist FPR against the probabilities obtained empirically from our four null artificial datasets. For all four datasets [OLS] does, as expected, produce a log probability plot that matches the nominal/theoretical frequentist FPR. However, this is not true for the [BIDET] approach.
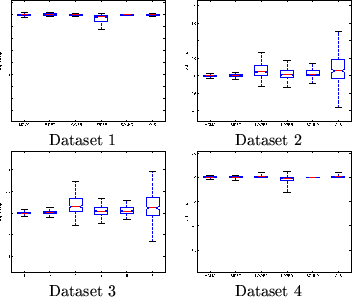
Datasets 1 and 4 with small
![]() compared to
compared to
![]() gives close to the same inference using [BIDET] as when
using [OLS]. Hence, we would expect the log probability that
[BIDET] produces to match the nominal/theoretical frequentist FPR.
Figure 6 demonstrates that this is true.
gives close to the same inference using [BIDET] as when
using [OLS]. Hence, we would expect the log probability that
[BIDET] produces to match the nominal/theoretical frequentist FPR.
Figure 6 demonstrates that this is true.
However, for Datasets 2 and 3 (
![]() is of the same
order as
is of the same
order as ![]() ) [BIDET] produces different results to [OLS].
The empirical log probabilities are lower than the
nominal/theoretical FPR in figure 6(b) and (c).
Recall from section 6.1.3, that we have two
ways in which we expect z-statistics to change between [OLS] and
[MCMC]. Firstly, they can increase due to increased efficiency
from using lower-level variance heterogeneity. Secondly, they can
decrease due to the higher-level variance being constrained to be
positive. The first of these effects will introduce no bias into the p-p plots. Hence, only the second of these effects will be visible and the p-p plots for
datasets 2 and 3 in figure 6 are consistent with this.
) [BIDET] produces different results to [OLS].
The empirical log probabilities are lower than the
nominal/theoretical FPR in figure 6(b) and (c).
Recall from section 6.1.3, that we have two
ways in which we expect z-statistics to change between [OLS] and
[MCMC]. Firstly, they can increase due to increased efficiency
from using lower-level variance heterogeneity. Secondly, they can
decrease due to the higher-level variance being constrained to be
positive. The first of these effects will introduce no bias into the p-p plots. Hence, only the second of these effects will be visible and the p-p plots for
datasets 2 and 3 in figure 6 are consistent with this.
This means that whilst we produce more accurate estimates of the total mixed effects variance, it also means that the z-statistics resulting from [BIDET] are not standardised Normally distributed under the null hypothesis. This is not a problem if we just report posterior probability maps or use mixture modelling.
However, if we do choose to proceed with assuming that the z-statistics from [BIDET] are standardised Normally distributed, since the empirical log probabilities are lower than the nominal/theoretical frequentist FPR, then the validity of our statistics will not be violated. In other words, the z-statistics from [BIDET] are, on average conservative. The disadvantage of this is that we will lose some sensitivity when compared with using the unknown, correct null distribution. The advantage is that we can utilise cluster based inference techniques on the z-statistic maps, such as Gaussian Random Field Theory (21,26).
 |
 |
 |
 |
 |