

Next: Multi-Session FMRI data
Up: Experimental Methods
Previous: Experimental Methods
We acquired whole brain volumes (
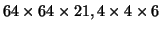 mm) of FMRI data on a Varian 3T system (TR=3sec; TE=30ms)
under resting condition. The data were corrected for subject motion
using MCFLIRT [Jenkinson et al., 2002], temporally high-pass filtered (Gaussian-weighted
least-squares straight line fitting, with
sigma=20.0s) [Marchini and Ripley, 2000] and masked for non-brain voxels using BET [Smith, 2002]. The pre-processed data was used to
estimate background noise parameters (voxel-wise means and std. deviations) which were used to generate 3 artificial data sets with
Gaussian noise characteristics. Artificial signal was
linearly added to the Gaussian background noise data using spatial maps and
time courses depicted in figure 1
mm) of FMRI data on a Varian 3T system (TR=3sec; TE=30ms)
under resting condition. The data were corrected for subject motion
using MCFLIRT [Jenkinson et al., 2002], temporally high-pass filtered (Gaussian-weighted
least-squares straight line fitting, with
sigma=20.0s) [Marchini and Ripley, 2000] and masked for non-brain voxels using BET [Smith, 2002]. The pre-processed data was used to
estimate background noise parameters (voxel-wise means and std. deviations) which were used to generate 3 artificial data sets with
Gaussian noise characteristics. Artificial signal was
linearly added to the Gaussian background noise data using spatial maps and
time courses depicted in figure 1![[*]](icons/crossref.png) . The time courses
correspond to the stimulus trains from a simple block design, a
single-event (fixed inter-stimulus interval) design and a single-event
(random inter-stimulus interval) convolved with a canonical
hæmodynamic response function (Gamma variate with 3s
standard-deviation and 6s lag).
. The time courses
correspond to the stimulus trains from a simple block design, a
single-event (fixed inter-stimulus interval) design and a single-event
(random inter-stimulus interval) convolved with a canonical
hæmodynamic response function (Gamma variate with 3s
standard-deviation and 6s lag).
Five different data sets (
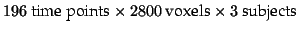 each) were generated as
example FMRI studies with different signal characteristics:
each) were generated as
example FMRI studies with different signal characteristics:
- (A)
- Each subject's data
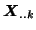 contains all three spatial maps
shown in figure 1
contains all three spatial maps
shown in figure 1![[*]](icons/crossref.png) .
Each spatial map has a different associated time course: time course 1
modulates spatial map 1, time course 2 modulates spatial map 2 and
time course 3 modulates spatial map 3. This defines
three spatio-temporal processes which are introduced at different
strengths into the individual subjects' data. The 'activation' levels were set to (3,4,5),(2,3,4) and (2,2,3) times the mean noise standard deviation for subjects 1-3. The complete 3-way data
conforms to the generative model of equation 1
.
Each spatial map has a different associated time course: time course 1
modulates spatial map 1, time course 2 modulates spatial map 2 and
time course 3 modulates spatial map 3. This defines
three spatio-temporal processes which are introduced at different
strengths into the individual subjects' data. The 'activation' levels were set to (3,4,5),(2,3,4) and (2,2,3) times the mean noise standard deviation for subjects 1-3. The complete 3-way data
conforms to the generative model of equation 1![[*]](icons/crossref.png) with
with 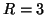 source processes in the data.
source processes in the data.
- (B)
- Each subject contains spatial map 1 modulated by time-course
1. In addition, subject 2 contains spatial map 2 modulated by time
course 2, while subject 3 contains spatial map 3 modulated by time
course 3. This data set is a special case of data set (A) with strength set to (3,0,0),(2,3,0) and (2,0,3). The data still conforms to the generative model of
equation 1
![[*]](icons/crossref.png) and is used to demonstrate the performance of
PARAFAC and tensor-PICA on data where the matric
and is used to demonstrate the performance of
PARAFAC and tensor-PICA on data where the matric 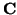 is sparse, i.e. for data which contains subject-specific source
processes in addition to a common source process.
is sparse, i.e. for data which contains subject-specific source
processes in addition to a common source process.
- (C)
- Like data set (A), but with the individual convolution
parameters for the generation of the signal time-courses differing
between subjects in mean lag and standard deviation used for the Gamma
HRF (
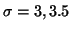 and
and 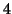 seconds, mean lag of
seconds, mean lag of 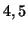 and
and 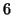 seconds). This induces small differences in the temporal signal
characteristics between
subjects. This data set is used to test for robustness against small
deviations from the model assumptions in the temporal domain (e.g. small differences between subjects in the BOLD response to the same
set of stimuli). Note that this data set still conforms to
the tri-linear model, as these different time courses together with the
spatial maps can be interpreted as separate source processes
(i.e. with
seconds). This induces small differences in the temporal signal
characteristics between
subjects. This data set is used to test for robustness against small
deviations from the model assumptions in the temporal domain (e.g. small differences between subjects in the BOLD response to the same
set of stimuli). Note that this data set still conforms to
the tri-linear model, as these different time courses together with the
spatial maps can be interpreted as separate source processes
(i.e. with
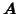 containing 9 time courses with sets of 3 time
courses being close to collinear and with
containing 9 time courses with sets of 3 time
courses being close to collinear and with
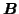 containing 9 spatial
maps where sets of 3 are identical). The data does not, however,
conform to the tensor-PICA model, as the spatial maps are not
statistically independent.
containing 9 spatial
maps where sets of 3 are identical). The data does not, however,
conform to the tensor-PICA model, as the spatial maps are not
statistically independent.
- (D)
- Subject 1 does not contain any 'activation' signal. Subjects 2 and 3 contain 'activation' signal in the area defined
by spatial map 2, modulated by the simple block-design (time course 1). Subject 3 also
contains extra 'activation' signal in the area defined by spatial map
3, modulated again by time course 1.
In addition, all three subjects contain 'nuisance'
signals (spatial map 1 modulated by a different time course in each
subject). This data simulates cases where FMRI data is confounded by e.g. resting-state networks which are spatially consistent but differ in
the temporal characteristics of the resting-state BOLD signal. The
data conforms to the tri-linear model when viewed as a set of 5
spatial maps with 5 associated time courses.
- (E)
- Each data set contains all three spatial maps, but modulated
by a different time course, i.e. the association between the
spatial maps 1-3 and time courses 1-3 changes between subjects. The
data conforms to the tri-linear model when viewed as a set of 9
spatial maps and 9 associated time courses. However, like data set (C) some
of the spatial maps are identical and thus not statistically independent.
Figure 1:
Artificial spatial maps and time-courses
used for the generation of artificial group data.
|
|


Next: Multi-Session FMRI data
Up: Experimental Methods
Previous: Experimental Methods
Christian Beckmann
2004-12-14
![]() each) were generated as
example FMRI studies with different signal characteristics:
each) were generated as
example FMRI studies with different signal characteristics:
![\includegraphics[width=\figwidth]{fig1}](img126.png)