Example Box: Registration
Introduction
The aim of this example is to become familiar with the role of cost functions and spatial transformations in registration.
This example is based on tools available in FSL, and the file names and instructions are specific to FSL. However, similar analyses can be performed using other neuroimaging software packages.
Please download the dataset for this example here:
Data download
The dataset you downloaded contains the following:
- Original (non-brain-extracted) T1-weighted image:
T1.nii.gz - Original (non-brain-extracted) T2-weighted image:
T2.nii.gz - Brain extracted versions of the above images:
T1_brain.nii.gzandT2_brain.nii.gz
Viewing the Data
To start with load both the original images (T1.nii.gz and T2.nii.gz) into a viewer (e.g. fsleyes) and inspect them. Note that they are not well aligned. Then load the brain extracted images (T1_brain.nii.gz and T2_brain.nii.gz) and check the quality of the brain extractions. Note that although the extractions are not perfect they are sufficiently good for registration purposes.
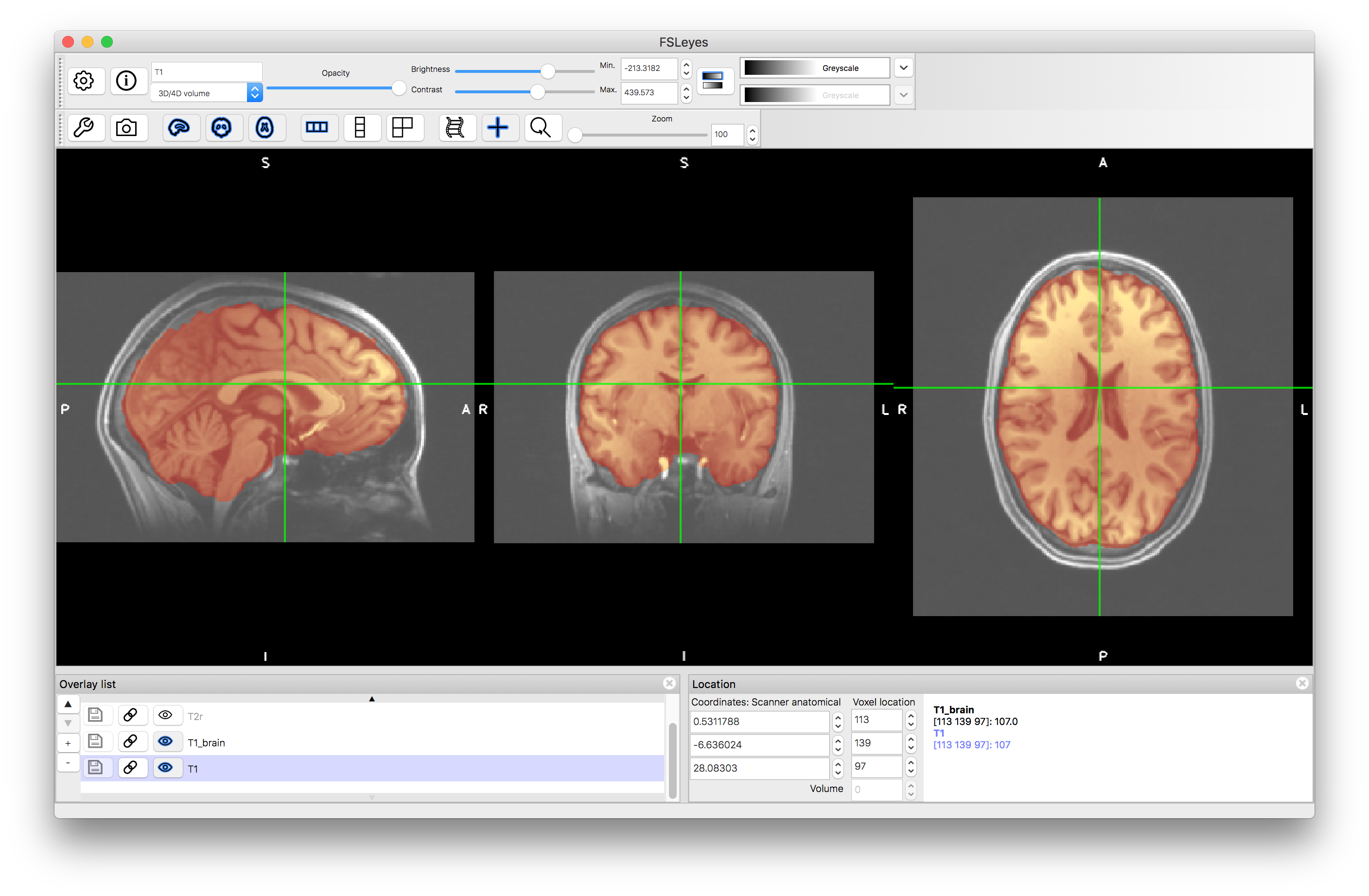
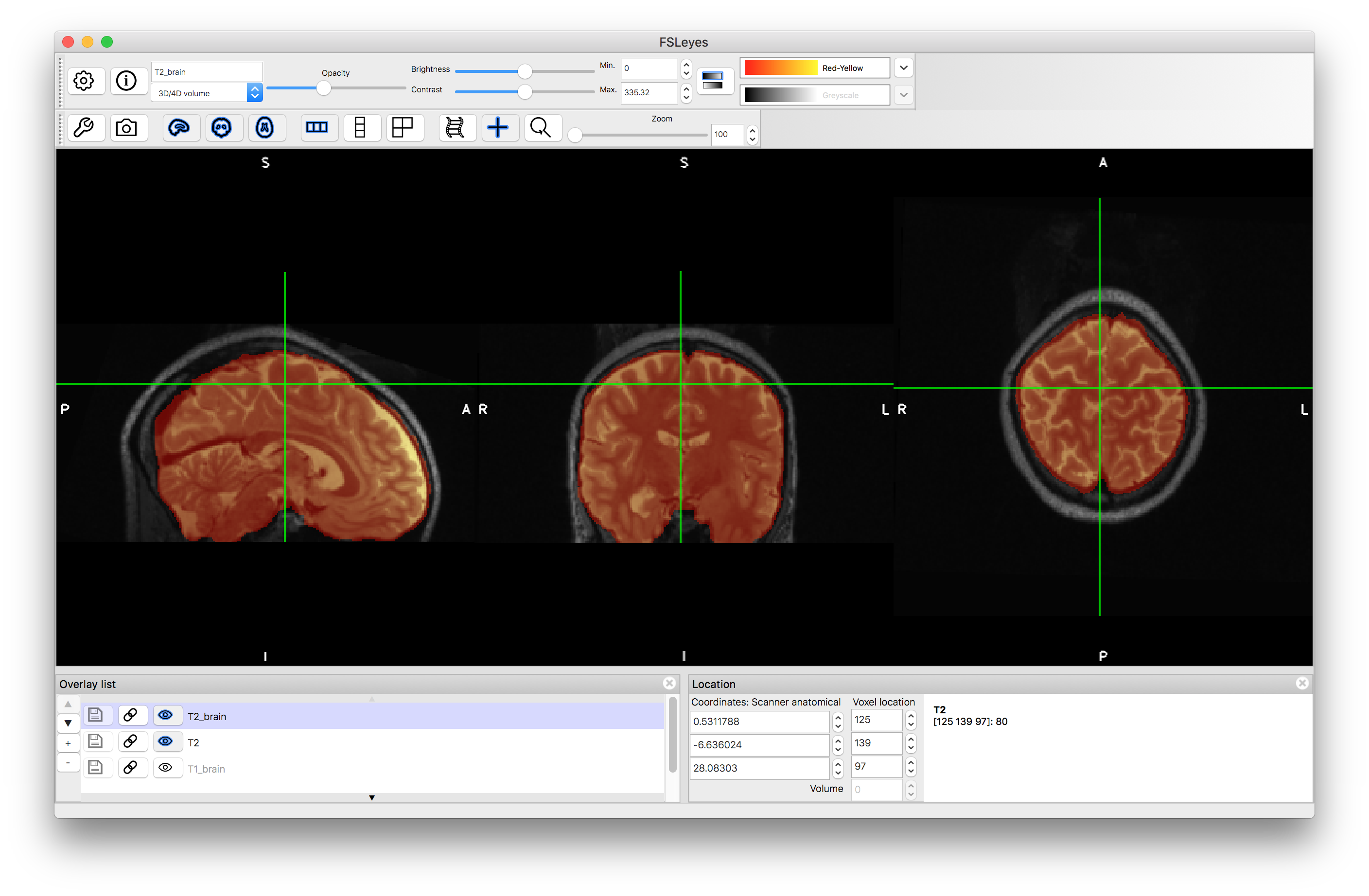
Within-subject Registration
The images here are from the same subject and hence should be registered using 6 DOF (rigid-body) since it is within-subject (we will register the T2-weighted image to the T1-weighted image). Since the images are different modalities then they should be registered using appropriate cost functions, but in order to demonstrate why this is we will try a range of cost functions. In FSL, the linear registration tool has a range of cost functions, and we will try five of them using the following commands:
flirt -in T2_brain.nii.gz -ref T1_brain.nii.gz -dof 6 -cost leastsq -omat T2toT1_LS.mat -out T2toT1_LS
flirt -in T2_brain.nii.gz -ref T1_brain.nii.gz -dof 6 -cost normcorr -omat T2toT1_NC.mat -out T2toT1_NC
flirt -in T2_brain.nii.gz -ref T1_brain.nii.gz -dof 6 -cost corratio -omat T2toT1_CR.mat -out T2toT1_CR
flirt -in T2_brain.nii.gz -ref T1_brain.nii.gz -dof 6 -cost mutualinfo -omat T2toT1_MI.mat -out T2toT1_MI
flirt -in T2_brain.nii.gz -ref T1_brain.nii.gz -dof 6 -cost normmi -omat T2toT1_NMI.mat -out T2toT1_NMI
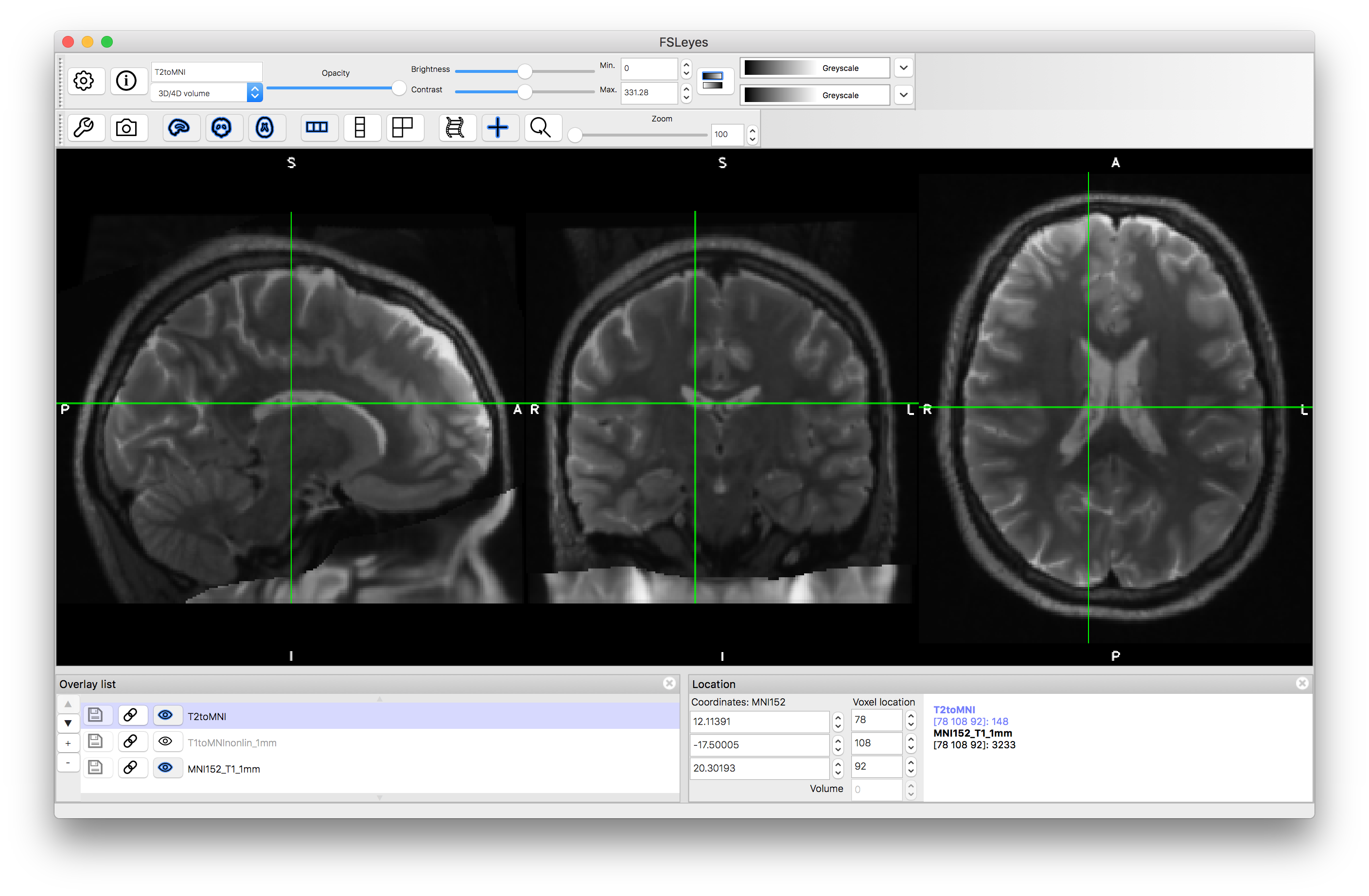
Once you have run the above commands, load the output images (e.g. T2toT1_LS.nii.gz) into the viewer so that you can assess the alignment with the T1-weighted image. To do this you can either: (i) turn all the images "off" (with the "eye" icon) except the T1 image and then turn "on" individual outputs and assess the alignment; or (ii) turn on a separate "ortho view" (via the View menu) and look at the T1-weighted image in one view and one of the outputs in the other - seeing if the crosshairs highlight the same anatomical location or not (as shown in the image below).
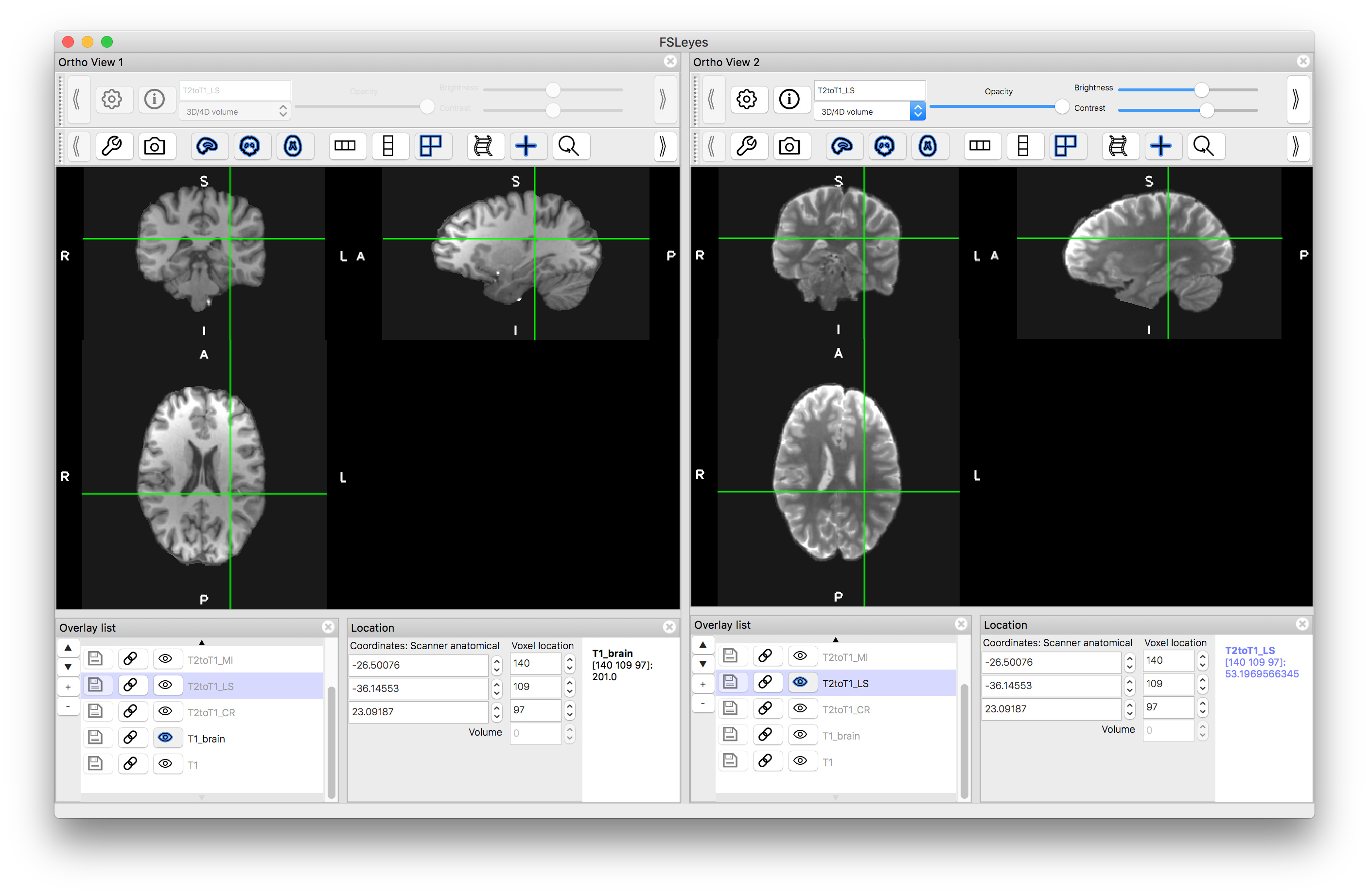
You should find that the alignment with the Least Squares and Normalised Correlation are the worst, as these are not suitable cost functions for registering images of different modalities. The other results should be clearly better, with the two mutual-information-based functions performing the best. This is partly due to the strong bias field that is in the images, since mutual information usually performs better in those circumstances. Removing the bias field as a pre-processing can considerably improve the result from Correlation Ratio, although will not have that much impact on the results from Least Squares or Normalised Correlation.
Also try running a 12 DOF registration using what you have decided was the best cost function from the above results. To do this just re-run the appropriate command from above, replacing -dof 6 with -dof 12 and altering the output filenames (e.g. -out T2toT1_MI_12 -omat T2toT1_MI_12.mat) to avoid it overwriting the previous results. Load the result in and see what the difference is like and whether it has made things obviously better or worse (it should be worse, as it is able to applying scalings and skews which are not needed, but can be driven by things such as noise or the differences in brain extraction, rather than by the true anatomical boundaries).
Subject to Standard Space Registration
The nonlinear registration tool in FSL (called FNIRT) can only register images of the same modality. Hence we will need to register the T1-weighted image to the T1-weighted MNI template (MNI152_T1). In order to run the nonlinear registration successfully we also need to initialise it by a linear (12 DOF) registration, to get the orientation and size of the image close enough for the nonlinear registration. Furthermore, the nonlinear registration does not use the brain extracted version, but the original (non-brain-extracted) image, so that any errors in brain extraction do not influence the local registration, whereas the linear registration does use the brain extracted versions (as was done above).
To register the T1-weighted image to standard space requires the following commands to be run:
flirt -in T1_brain.nii.gz -ref $FSLDIR/data/standard/MNI152_T1_2mm_brain.nii.gz -dof 12 -out T1toMNIlin -omat T1toMNIlin.mat
fnirt --in=T1.nii.gz --aff=T1toMNIlin.mat --config=T1_2_MNI152_2mm.cnf --iout=T1toMNInonlin --cout=T1toMNI_coef --fout=T1toMNI_warp
The outputs of the nonlinear registration (FNIRT) are: T1toMNInonlin.nii.gz which is the transformed image and two versions of the spatial transformation T1toMNI_coef and T1toMNI_warp. See the previous practical for more information about the spatial transformation files.
Load the nonlinear registration output T1toMNInonlin.nii.gz and compare the alignment to the standard image MNI152_T1_2mm. In addition, load the linear registration output (T1toMNIlin.nii.gz) and compare the accuracy of this alignment to the MNI template with the nonlinear output. You can also create a higher resolution version using this command:
applywarp -i T1.nii.gz -r $FSLDIR/data/standard/MNI152_T1_1mm.nii.gz -w T1toMNI_warp.nii.gz -o T1toMNInonlin_1mm.nii.gz
Load in the 1mm version of this and assess the alignment to the MNI152_T1_1mm standard template. You should find that major structures (e.g. ventricles) are very well aligned, although smaller individual folds will differ.
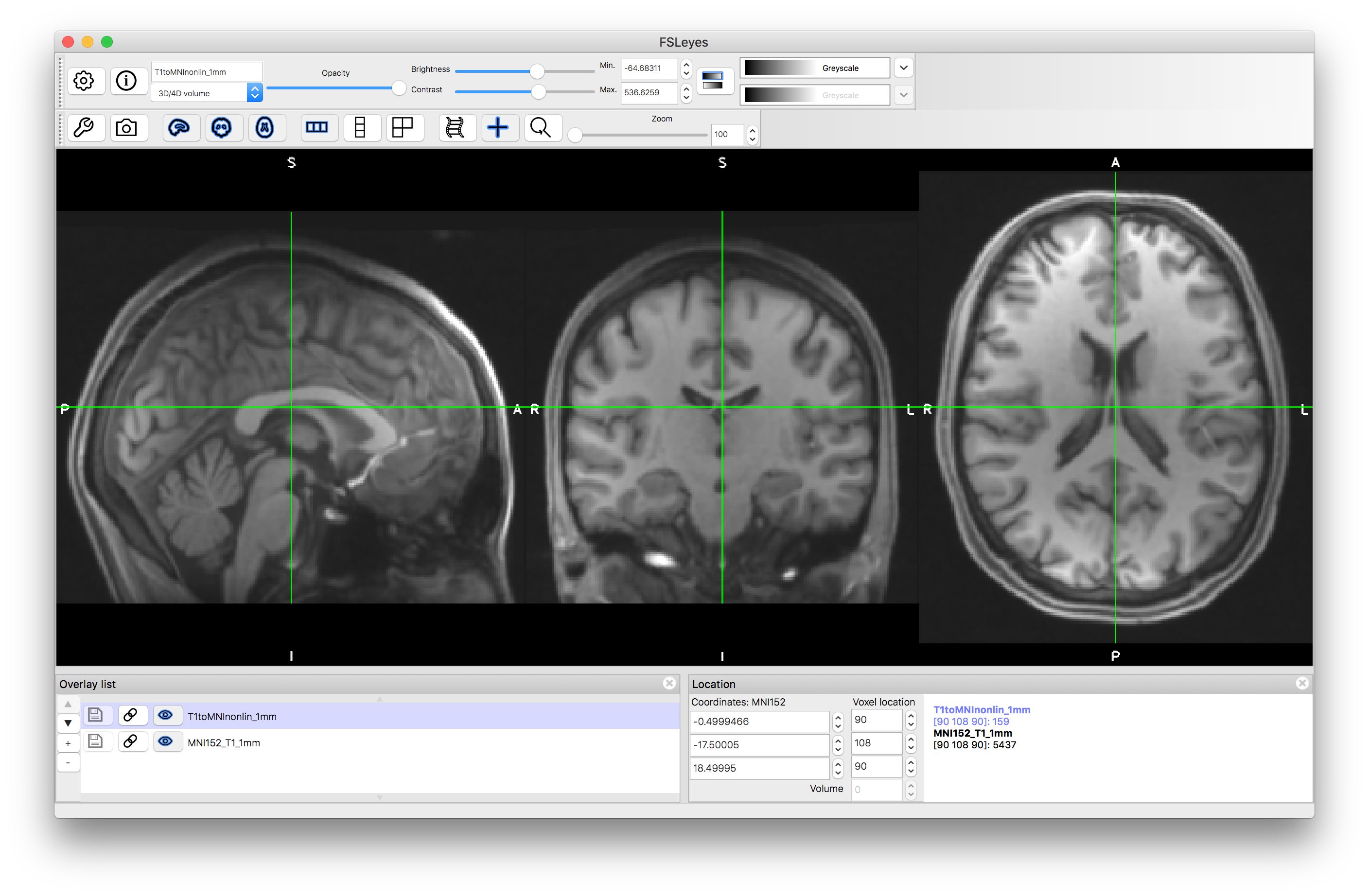
Combining Transformations
Finally, we will also transform the T2-weighted image into the MNI standard space. To do this we will combine the two transformations together: (1) the T2-weighted image to the T1-weighted image (within subject); and (2) the T1-weighted image to the MNI standard template. This combination (or concatenation) can be done with a single command:
applywarp -i T2.nii.gz -r $FSLDIR/data/standard/MNI152_T1_1mm.nii.gz --premat=T2toT1_MI.mat -w T1toMNI_warp.nii.gz -o T2toMNI.nii.gz
Once you have run this command, load the output (T2toMNI.nii.gz) on top of the MNI template, along with the T1toMNInonlin_1mm.nii.gz result. Inspect the results: the T2-weighted and T1-weighted images should be very well registered together, and these should be aligned well with the main features of the MNI template (e.g. subcortical structures, major sulci) but will be less accurately aligned for minor folds.
From these examples you should gain an understanding of what cost functions should be used, and how they affect the registration results, as well as how to combine different transformations, with different degrees of freedom.