Example Box: Distortion correction with blip-up-blip-down data
Introduction
The aim of this example is to become familiar with how to use blip-up-blip-down image pairs (also known as phase-encode-reversed pairs) to perform distortion correction and registration of dMRI (or fMRI) and structural images.
This example is based on tools available in FSL, and the file names and instructions are specific to FSL. However, similar analyses can be performed using other neuroimaging software packages.
Please download the dataset for this example here:
Data download
The dataset you downloaded contains the following:
- Original (non-brain-extracted) T1-weighted image:
struct.nii.gz - A single diffusion-weighted volume from the main diffusion acquisition:
diffvol.nii.gz - A single non-diffusion-weighted (b=0) volume from the main diffusion acquisition:
nodif.nii.gz - A single non-diffusion-weighted (b=0) volume with opposite phase-encoding direction:
nodif_PA.nii.gz - A text file containing some key acquisition parameters:
acqparams.txt - Directory containing previously generated results:
pre_baked
Viewing the data
Start by loading the two b=0 images (those without diffusion weighting) with opposite phase encoding directions (nodif.nii.gz and nodif_PA.nii.gz) into a viewer. The main phase-encoding direction for this dataset is AP (anterior to posterior) and the nodif.nii.gz image is extracted from the main acquisition. The other image is a separate acquisition (nodif_PA.nii.gz), with no diffusion weighting and a reversed phase encoding direction (PA = posterior to anterior). For this example we only have one 3D volume in each case, but in practice we recommend obtaining several (e.g. 3-5 phase-encode reversed b=0 images plus the many b=0 images that should be acquired with the main diffusion dataset).
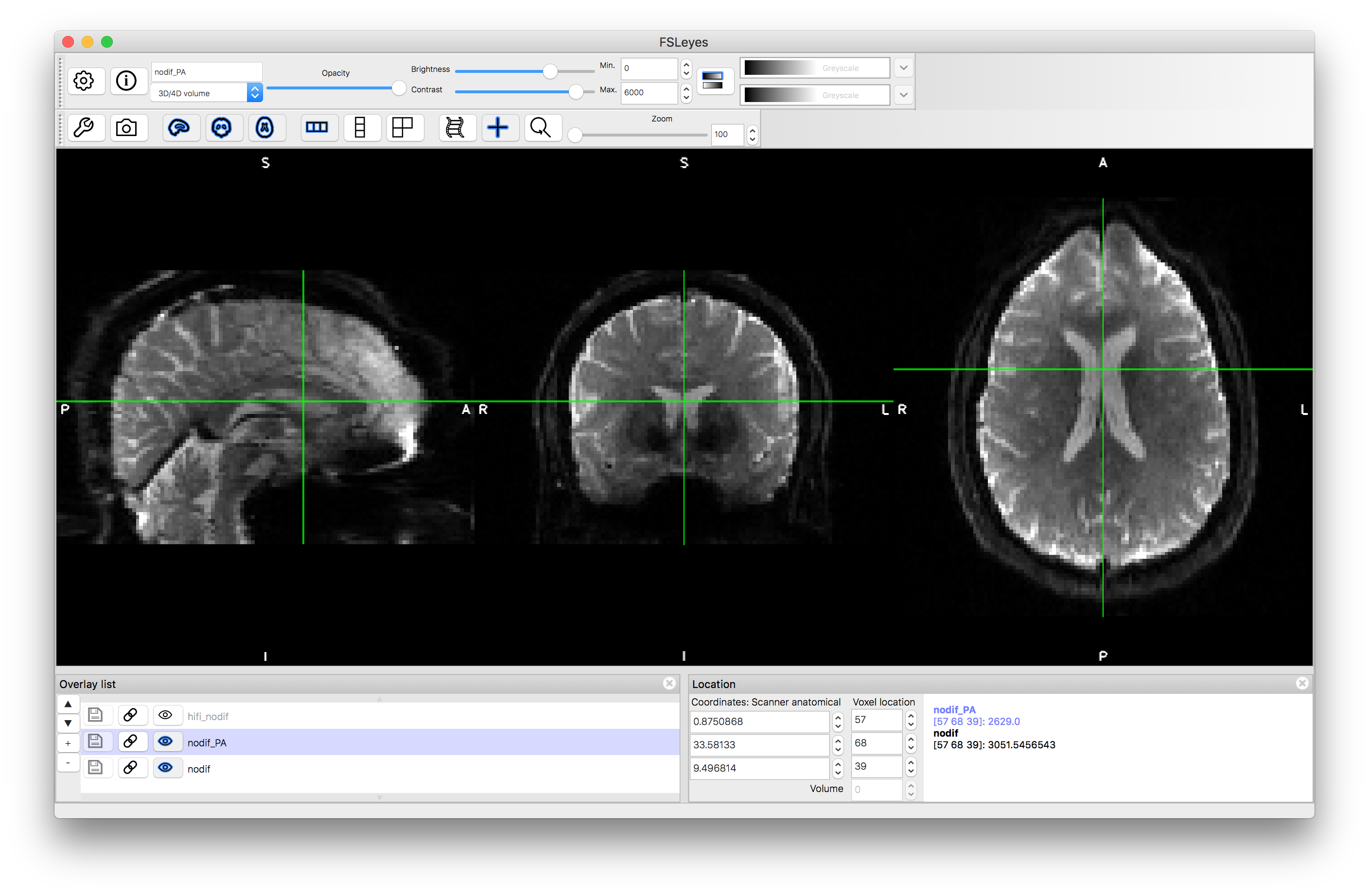
Flick between the two images and see how the geometric distortion is reversed. Also notice how the intensity is brighter around rims where the distortion has compressed the signal (e.g. in the anterior portion of the AP image, shown in the image below).
Phase encoding = AP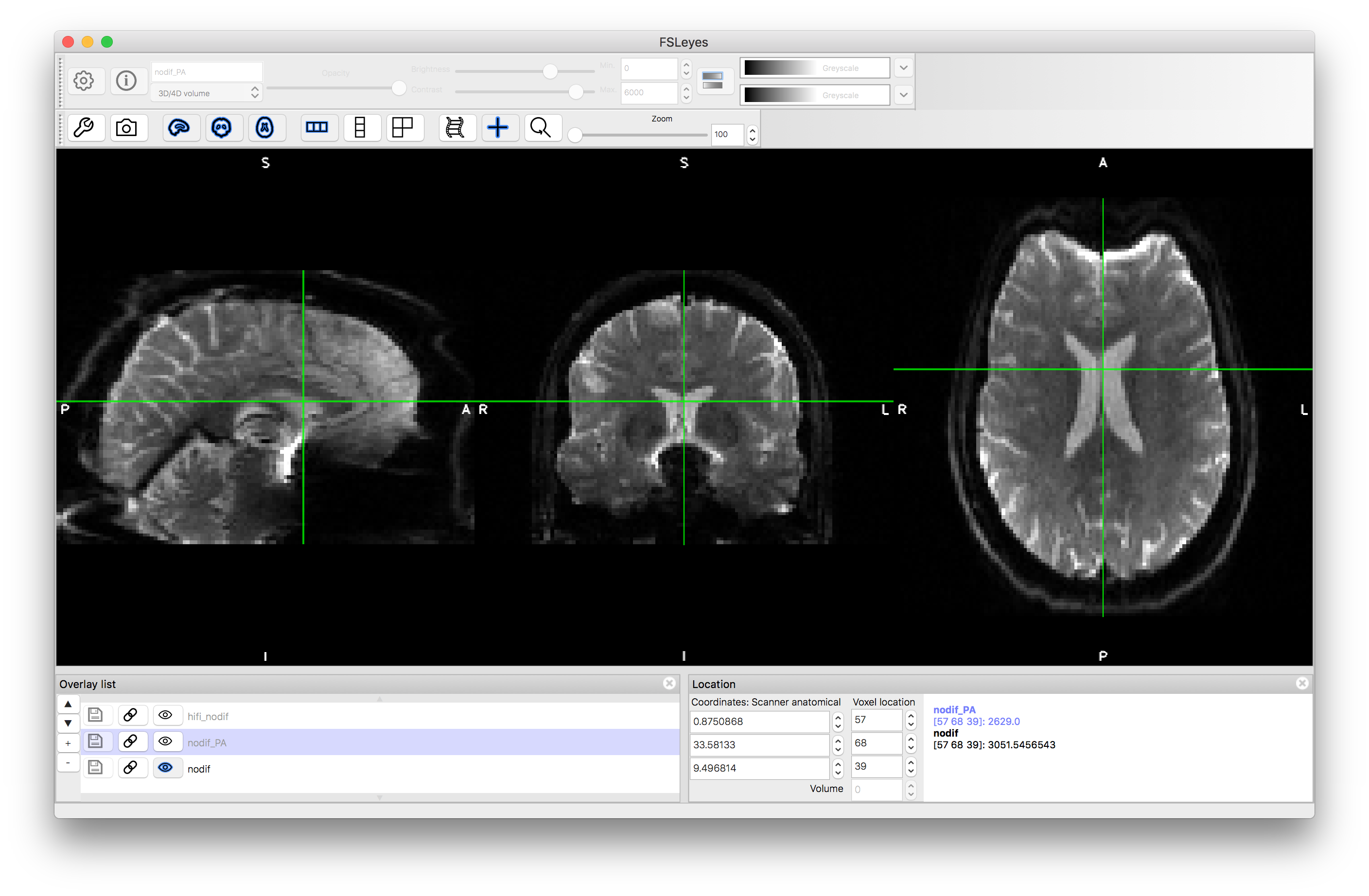
Phase encoding = PA
Data preparation
To run the FSL tool (topup) for distortion correction with blip-up-blip-down data we need to first merge the b=0 (non-diffusion-weighted) images together into a single 4D image. This can be done with the following command:
fslmerge -t AP_PA_b0.nii.gz nodif.nii.gz nodif_PA.nii.gz
The order of the images in this file is important and needs to match with the order in the acqparams.txt file. It is this file that contains information about the phase encoding directions and the "total readout time" for each image (the total readout time is the amount of time between the centre of the first and last echoes in k-space - it is not important to understand what this means, as your scanner operator will be able to give you this value). The values in the acqparams.txt file are:
0 -1 0 0.0665 0 1 0 0.0665
The first three columns specifies the direction of the phase encoding. The non-zero number in the second column means that is is along the y-direction, with a -1 meaning that k-space was traversed in the Anterior to Posterior direction and a +1 meaning that it was traversed in the Posterior to Anterior direction. The final column specifies the "total readout time", in seconds. The first row corresponds to the first volume in the 4D merged image (nodif.nii.gz) and the second image corresponds to the second volume in the 4D image (nodif_PA.nii.gz).
Distortion correction
Given the above files we can now run the distortion correction using the following command:
topup --imain=AP_PA_b0 --datain=acqparams.txt --config=b02b0.cnf --out=topup_AP_PA_b0
There are two result files, one called topup_AP_PA_b0_fieldcoef.nii.gz which contains information about the B0 field and topup_AP_PA_b0_movpar.txt which contains information about any movement between nodif.nii.gz and nodif_PA.nii.gz. The topup_AP_PA_b0_fieldcoef.nii.gz file can be viewed with fsleyes. It looks like a low resolution fieldmap, and it contains the spline coefficients for the field that topup has estimated. To save the actual field it is necessary to add --fout=my_field to the topup command line, but we will not need that here.
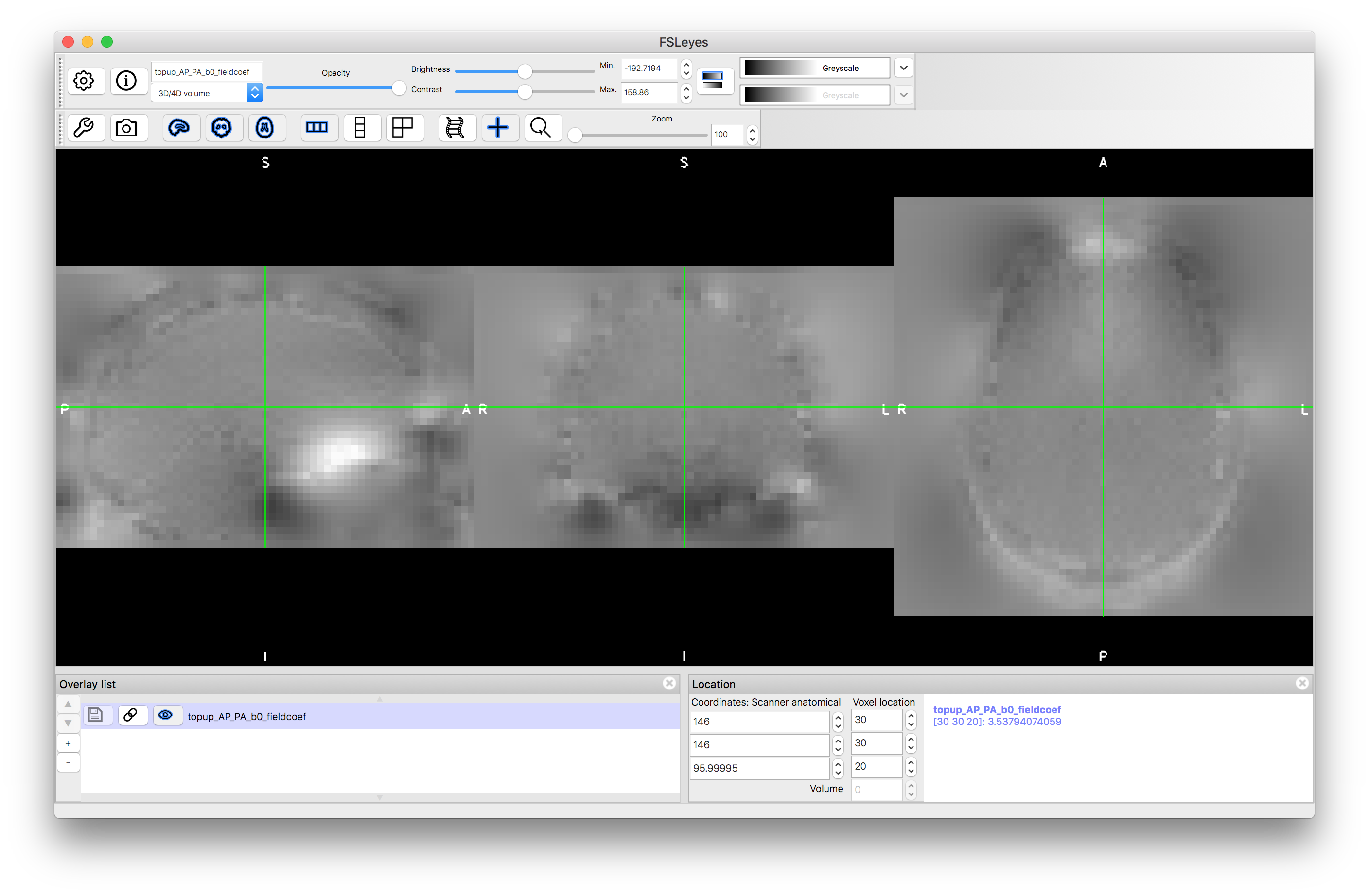
To use the field output by topup to resample an image and correct the distortion, a separate tool - applytopup - needs to be run. It is separated here because normally the topup field information is combined with other information, such as eddy-current corrections, to simultaneously correct for multiple distortions. In our case we will just correct the non-diffusion-weighted images, which is sufficient for registration. The command for distortion correction in this case is:
applytopup --imain=nodif,nodif_PA --topup=topup_AP_PA_b0 --datain=acqparams.txt --inindex=1,2 --out=hifi_nodif
This will generate a file named hifi_nodif.nii.gz where both input images have been combined into a single distortion-corrected image. Load this into a viewer and compare it to nodif.nii.gz and nodif_PA.nii.gz - you will see that it appears practically distortion free.
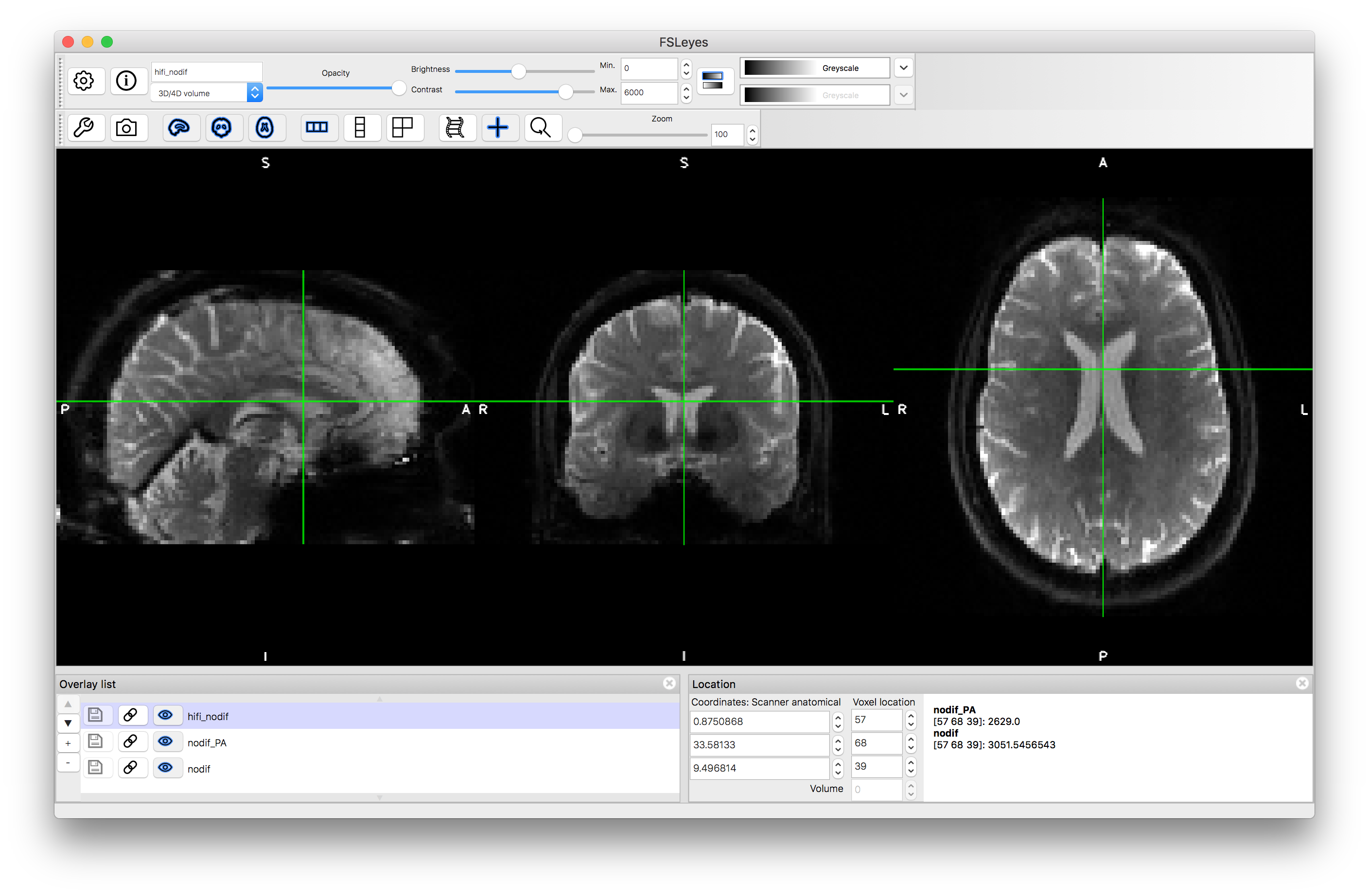
Registration
The distortion-corrected diffusion image can be aligned to the structural image using a 6 DOF (within-subject) registration with BBR. Since the distortion has already been corrected, no extra fieldmap information is needed and we can make a simpler call to the epi_reg script than in the previous example box that used gradient-echo fieldmaps.
The commands needed are:
bet struct.nii.gz struct_brain.nii.gz
epi_reg --epi=hifi_nodif.nii.gz --t1=struct.nii.gz --t1brain=struct_brain.nii.gz --out=diff2struct
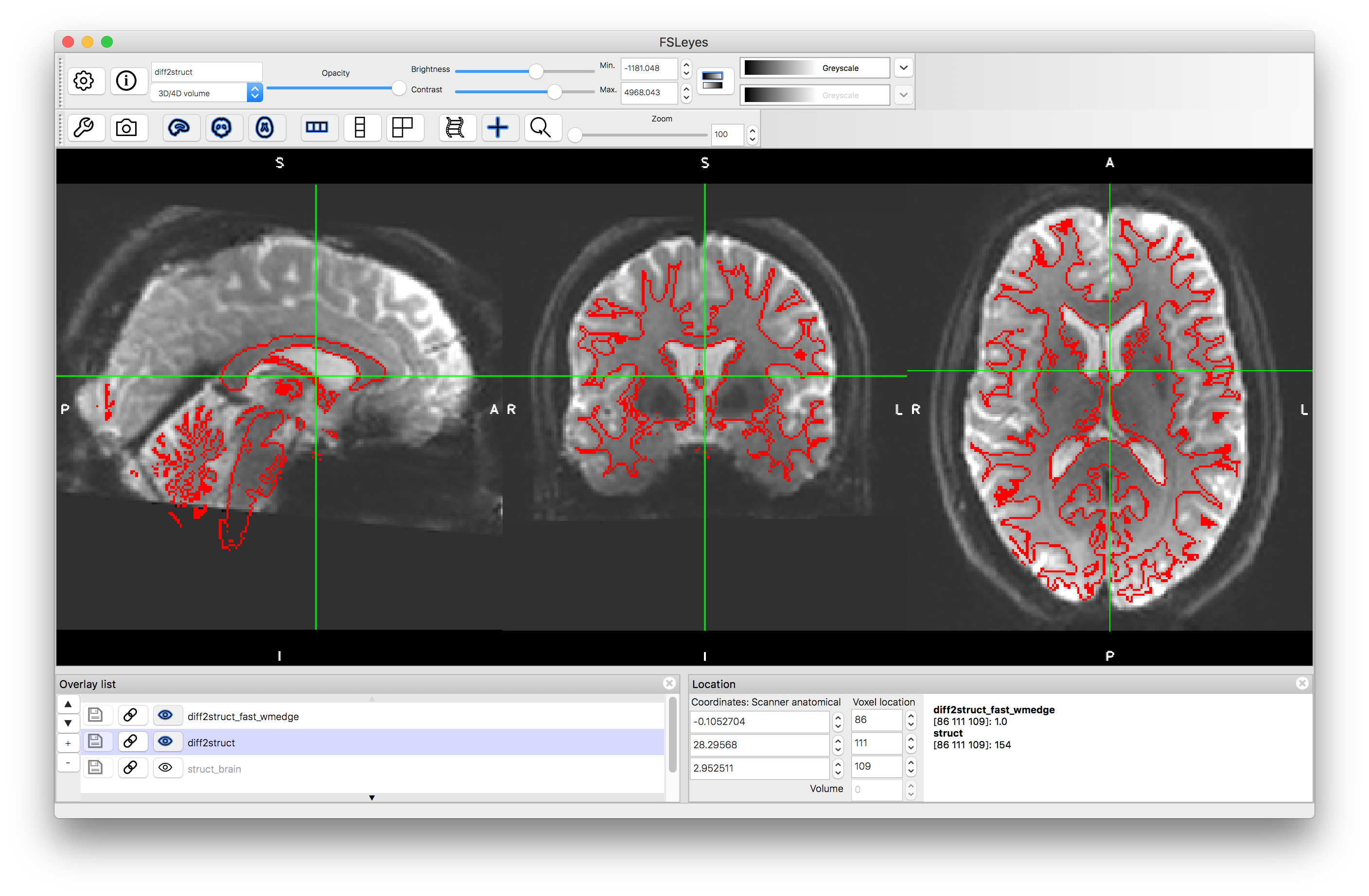
The main outputs of epi_reg are a resampled version of the diffusion image in structural space (diff2struct.nii.gz) and the spatial transformation corresponding to this (diff2struct.mat). There is also the white matter boundary in diff2struct_fast_wmedge.nii.gz.
Load the transformed diffusion image (diff2struct.nii.gz) along with the structural image (struct.nii.gz) and the white matter boundaries (diff2struct_fast_wmedge.nii.gz) - giving the latter a Red colourmap. Viewing the boundaries over the transformed diffusion image should show you how accurate the alignment is (as can flicking between the diffusion and structural images).
If other images need to be transformed or distortion corrected then this can be achieved by combining transformations together (the 6 DOF one from registration and the warp field from the distortion correction) and then applying them using tools such as applywarp or by using the fieldmap derived from topup as a substitute for a gradient-echo fieldmap. This allows any images to be corrected; that is, not only diffusion images but also functional images.
From these examples you should be able to prepare data and run distortion correction and registration using blip-up-blip-down images. This is not the only way to perform distortion correction, but is the most advantageous when working with dMRI data or both dMRI and fMRI data. The alternative for distortion correction is to use gradient-echo fieldmaps, which is covered in the previous example box and can also be applied to either fMRI or dMRI data (although for dMRI data there are advantages in using blip-up-blip-down images).